Workflow
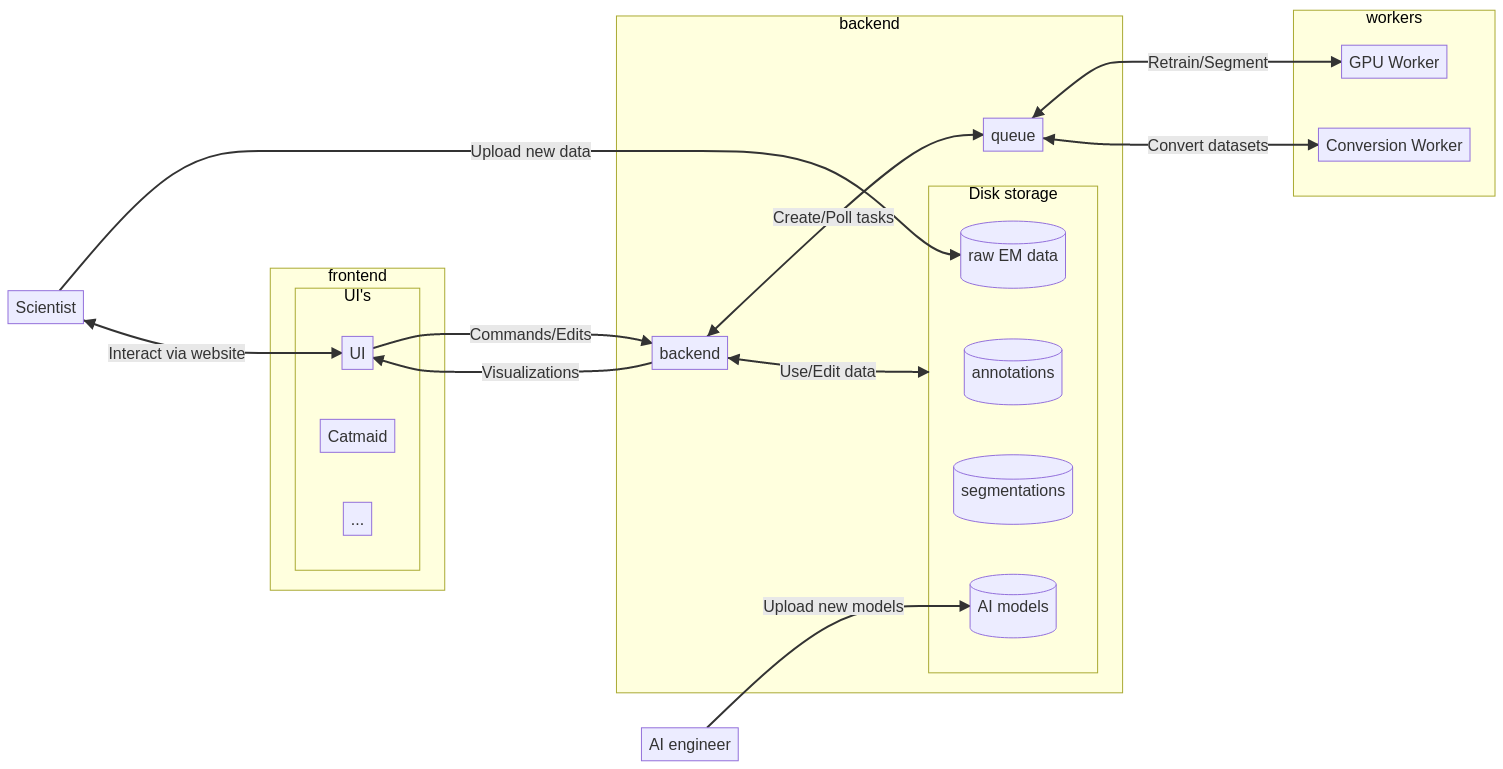
The following diagram gives on overview of the BioSegment workflow. Users interact with a frontend using their browser. They can visualize a dataset, edit annotations and create segmentations using AI models. The BioSegment backend handles the tasks given by the frontend and fetches the datasets from disk storage. For long-running tasks like conversion and fine-tuning, separate workers are used.
Data¶
- Dataset
- Electron-microscopy data
- example formats: pngseq, tif3d
- Classes of interest
- example classes of interest: mitochondria, endoplasmatic reticulum...
- Segmentations
- Attribution for each part of a dataset to an interest
- mostly ground-truth or machine made
- Annotations
- Stroke or area of a part of the dataset that is attributed
- made by a human
- AI models
- able to take EM data and an annotation and create a segmentation
- can be pretrained and further fine-tuned with additional annotations
- e.g. UNet
Actors¶
- Scientist
- A domain expert that wants to visualize and annotate EM data with a specialized tool
- AI engineer
- Implements and pretrains AI models
User flow¶
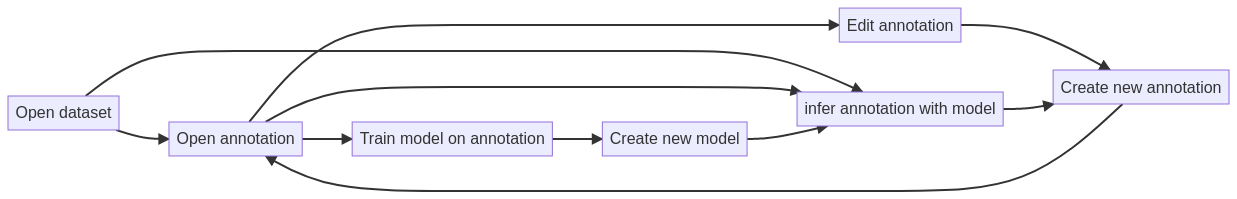
Example of the user flow for a scientist when interacting with a BioSegment frontend.